The Andersson lab aims to understand how specialized organs systems develop the correct cellular identity and architecture. As developmental biologists, we are interested in why and how genetic mutations lead to specific defects in embryogenesis. We develop new methods to analyze organ architecture, or new technologies to manipulate embryogenesis in mice, with emphasis on the 3Rs. We are focused on the Notch-related disorder Alagille syndrome, a congenital disorder affecting development of several organ systems including the liver and vasculature. In addition to the technologies we are developing, we use mouse models, 3D cell culture, CRISPR cell lines, single cell omics, and patient samples to address fundamental questions with relevance for human health.

Andersson Lab May 2022 (Photo by Linda Lindell). First row (sitting, left to right): Bettina Semsch, Elisabeth Verboven, Sandra De Haan, Noemi Van Hul, Katrin Mangold (alumnus, defended PhD Student). Second Row (standing): David Kosek, Afshan Iqbal, Lenka Belicova, Paulina Zydowicz-Machtel, Jia Sun, Emma R Andersson, Lukas Poppe, Jingyan He, Agustin Corbat
News
October 2024
Masek et al, EMBO Molecular Medicine, out now, and highlighted in KI News! Patients with Alagille syndrome have an atypical fibrotic response to liver disease. In this paper, we discovered and dissected apart the role of the dysregulated immune system and liver developmental delay in controlling the fibrotic response to cholestasis in a mouse model of Alagille syndrome. This is a project Dr Masek started as a postdoc in the Anderson lab, and then finalized in collaboration with the lab he established at Charles University in Prague.
September 2024
De Haan et al, accepted in Development, link coming soon! Notch signaling orchestrates the intricate patterning of the inner ear, precisely regulating the formation of sensory hair cells and supporting cells. Disruptions in this pathway, such as mutations in the Notch ligand JAG1 found in patients with Alagille syndrome, are associated with hearing impairments. Despite these clinical links, the role of Notch signaling in inner ear development had not been explored in a relevant disease model—until now. Using whole-mount imaging, single-cell RNA sequencing, and RNAscope, we investigated Jag1’s function at the cellular level in the developing organ of Corti. Our findings reveal a novel role for Jag1 as a repressor of Notch signaling in supporting cells, crucially inhibiting the formation of outer hair cells within the inner hair cell compartment. This discovery uncovers a new layer of complexity in inner ear patterning and offers fresh insights into the molecular underpinnings of hearing disorders associated with Notch signaling defects.
August 2024
August 30th: congratulations Sandra de Haan on the successful defense of your PhD thesis! Dr de Haan is the 6th PhD student graduating from the Andersson lab. We thank Amy Kiernan (Universty of Rochester, USA) for acting as expert Faculty opponent, as well as Sten Linnarsson, Pierre Hakizimana and Helge Rask-Andersen for participating in the Examination Board.

New publication! Hildebrandt et al: In collaboration with Johan Ankarklev at Stockholm University, we have contributed to resolving host-pathogen interactions in Plasmodium-infected mouse liver with spatial transcriptomics. Special thanks to Noemi Van Hul (our lab) and first author Franziska Hildebrandt (Ankarklev lab, former PhD student, now defended).
May 2024
Our News & Views “‘Understanding liver repair through space and time” in Nature Genetics (Belicova*, Van Hul* & Andersson, Nature Genetics 2024) is out! Written together with Lenka Belicova and Noemi Van Hul, we discuss two exciting new papers leveraging Stereo-Seq to resolve and map molecular signatures of liver cell types and their interactions in homeostasis, damage, repair and regeneration.
March 2024
Two fresh new PhDs defended in the Andersson Lab! Congratulations to:
Dr Afshan Iqbal, with the thesis “Mechanisms Driving Notch Disorders and Therapeutic Implications”. We (Afshan and Emma) thank her outstanding opponent Professor Rebecca Wells.
Dr David Kosek, with the thesis “The role of pairing beyond the seed in microRNA targeting. We (David, Emma, and Davids’ cosupervisor Prof Katja Petzold), thank his fantastic opponent Professor Francois Major.
February 2024
Two new publications from the Andersson Lab!
Iqbal et al, Liver International 2024: On the cover, at right. In this original publication, Afshan, Noemi, Lenka, Agus and Simona addressed whether biliary cells from the center vs the periphery of the liver are different in a model of Alagille syndrome, after the liver has regenerated its biliary system (a phenomenon that happens spontaneously in some mice and some people with Alagille syndrome!) Using organoids, liver histology, RNA sequencing and advanced image analyses, they showed that not only are central and peripheral bile ducts different from one another in disease, they are also both different from healthy bile ducts and react differently to a possible drug treatment (IGF1). Interestingly, bile duct organoids from the central region spontaneously developed hepatocyte cells, and in ALGS mice ectopic hepatocytes could be found embedded in portal vein mesenchyme next to bile ducts. We speculate that the biliary identity, even once established, is not well maintained Alagille syndrome.
Masek and Andersson, Current Opinion in Cell Biology 2024: Jan Masek, a former postdoc in the Andersson group – now a group leader at Charles University in Prague, and Emma wrote a review on how JAG1 controls embryonic development, and the consequences of dysregulated JAG1 signaling in Alagille syndrome.
December 2023
Congratulations Dr Jingyan He on the successful defense of your PhD thesis! Many thanks to the excellent examination board (Prof Goncalo Castelo Branco, Dr Fredrik Lanner and Dr Claudio Cantu), our distinguished opponent Professor Anna Mae Diehl, and our Chair Dr Maria Kasper.
October 2023
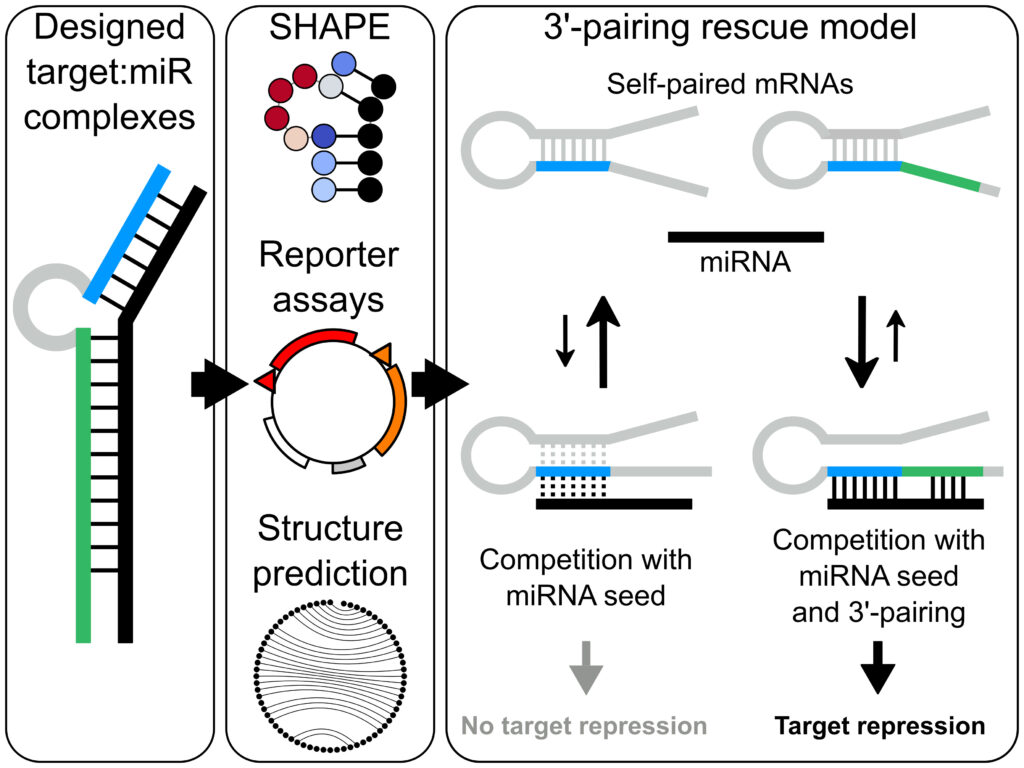
Congratulations David Kosek on the publication of your first first-author paper in Nucleic Acids Research! Understanding the rules of miRNA-mRNA target engagement and functional outcomes are essential for refinement of miRNAs or siRNAs as therapeutic tools. David, a PhD student in the Andersson lab who is part of a tight KAW-funded collaboration with the Petzold lab, has studied the impact of miR binding capacities on target downregulation. David uncovered a function for 3′ pairing by the miRNA in rendering mRNA target sites less sensitive to seed-site occlusion in the native mRNA structure. For more, check out the publication online here (Kosek et al, Nucleic Acids Research 2023)!
June 2023
Congratulations to our PhD student Afshan Iqbal, who was awarded the prize for Best Basic Science Poster at the EASL Congress!
Our lab submitted 4 abstracts to the upcoming EASL Congress, three of which were ranked in the Top 10% and competed for Best Poster Prizes (the fourth abstract was an update of an abstract that was previously top-ranked at an EASL congress). Afshan Iqbal, Dr. Elisabeth Verboven, Linde Sevenants & Dr. Noemi Van Hul (co-presenting a poster) and Jingyan He presented our latest insights into bile duct de novo growth in a model of Alagille syndrome using hilar or peripheral biliary organoids, developmental hepatic innervation processes, spatial transcriptomics of the atypical Alagille syndrome fibrotic response, and a new technique to manipulate hepatic stellate cells in utero.
March 2023
Banijamali et al published in RNA! In this first collaborative publication with the Petzold lab, we report the further development of selective 2′-hydroxyl acylation analyzed by primer extension (SHAPE) to resolve how microRNA-34a (miR-34a) binds its mRNA target, the silent information regulator 1 (mSIRT1), both with and without the Argonaute protein. This is the first paper from our collaboration that was seeded in 2016, in which we ultimately aim to resolve miR:mRNA selectivity principles in vivo, using our lab’s technique NEPTUNE. More papers in the pipeline, coming soon!
Dec 2022
Hankeova et al published in EMBO Molecular Medicine and featured on the cover! Simona Hankeova (former PhD student in the lab, now post doc at Genentech), addressed why patients with Alagille syndrome experience spontaneous bleeds, using our mouse model, patient retinographs, and a systematic literature review. With thanks to Noemi Van Hul and Eli Verboven who championed the revision process and ensured that the paper was finalised. Fantastic collaboration with clinicians, scientists worldwide, and over many years. Thank you to the patients for participating in the research process. And thank you to the constructive reviewers through the Review Commons process, an excellent system!
June 2022

The Sweden Liver Week – Highlight of Swedish Hepatology!
Emma gave the State of the Art Lecture on June 15th.
The Sweden Liver Week is ongoing and three labmembers were selected to give talks: congratulations to Jingyan He, Paulina Zydowicz-Machtel and Lukas Poppe! And a massive thank you to the co-organizers, which include our very own Senior Scientist Noemi Van Hul (abstract selection process was independent of Noemi 😉 )
Congratulations to Jingyan He, whose presentation was selected as Best Talk on Day 1!
And Congratulations to Lukas Poppe, Master’s student in our lab supervised by Noemi Van Hul, whose presentation was selected as Best Talk on Day 2!

Congratulations to Dr Katrin Mangold who successfully defended her PhD!
Dr Mangold is the second PhD graduate from our lab. Her work focused on developing NEPTUNE to target nervous system with high efficiency, and develop it for neural and ectoderm-derived tissues.
May 2022
Our review on “Modern therapeutic approaches to liver-related disorders“, led by Pavel Strnad (University Hospital RWTH Aachen, Germany) together with Antoine Gardin, Katharina Remih and Emmanuel Gonzales, is out in Journal of Hepatology! We discuss challenges and opportunities in siRNA, modified RNA, gene engineering etc to treat liver disease and diseases stemming from liver dysfunction.

April 2022
We welcome our new postdoc Agustin Corbat, who performed his PhD in the Quantum Electronics Lab under the direction of Dr. Hernan Grecco at the Department of Physics of the Exact and Natural Sciences Faculty of the University of Buenos Aires, Argentina.
March 2022

Lenka Belicova (left) and Elisabeth Verboven (right) both awarded Marie Curie fellowships in the EU Horizon MSCA Postdoctoral Fellowships 2021 call! Lenka will use barcode lineage tracing to map liver cell plasticity (Mangold et al, Cell Reports Methods 2021, Kameneva, Artemov et al Nature Genetics 2021) and Eli will investigate the role of the nervous system in liver health and disease. Thank you to the reviewers for seeing the potential in these projects!
Editorial by ER Andersson out now in Hepatology. This editorial discusses exciting new work from Duc Dong’s lab, in the context of previous findings from Stacey Huppert and Holger Willenbring’s labs.
February 2022
In order to disseminate neural plate targeting with in utero nano-injection (NEPTUNE) (Mangold et al Cell Reports Methods 2021), we are happy to present the full protocol in the Journal of Visualized Experiments (JoVE): Mangold et al, Jove 2022
Also, if you are interested in NEPTUNE or in utero genetic manipulation for your experiments, we set up a core facility to provide NEPTUNE as a service to researchers; INFINiGENE (INFrastructure for in utero GENEtic manipulation).
Our Special Issue of the European Journal of Medical Genetics “Advances in genetic, epigenetic and environmental aspects of rare liver diseases” is out. ER Andersson co-guest-edited this Special Collection together with Prof Dr Ansgar Lohse, on behalf of the European Reference Network for Rare Liver Disease (for which ER Andersson is the External Scientific Advisor). The collection highlights progress in rare diseases such as PSC, PFIC, Wilsons disease and more, and includes an editorial introduction that you can read here.
December 2021
Collaborative paper led by Franziska Hildebrandt in Johan Ankarklev’s lab at Stockholm University published in Nature Communications, describing the application of spatial transcriptomics to large liver tissue slices. We confirm liver zonation principles and demonstrate how this will be useful to further studies of liver architecture.
November 2021
Congratulations to Elisabeth Verboven for being awarded a David and Astrid Hagelen Foundation Fellowship for her postdoctoral project! This fellowship will support Dr Verboven for two years, and we are grateful to the foundation and reviewers for finding the project and Dr Verboven meritorious!
Congratulations to Sandra De Haan for being awarded two travel grants to present her work at the ARO 2022 MidWinter Meeting in California! Sandra will present her work on the inner ear development and homeostasis in a mouse model for Alagille syndrome.
Emma R Andersson is awarded a Karolinska Institutet Consolidator Grant! We thank both the internal and external reviewers!
October 2021
Andersson lab is awarded a 3R Grant from the Swedish Research Council to continue their work developing in utero nano-injection as an alternative method to traditional mouse genetics – aiming to reduce the numbers of animals needed in life sciences. Thank you for the continued support!
September 2021
In order to disseminate double resin casting microCT (DUCT) (Hankeova & Salplachta et al eLife 2021), we are happy to present the full protocol in the Journal of Visualized Experiments (JoVE): Hankeova & Salplachta et al JoVE 2021.
July 2021

On the cover: Our latest paper is out in Cell Reports Methods. We developed a technique to efficiently manipulate gene expression in the embryonic mouse nervous system. We called the technique NETPUNE for neural plate targeting by in utero nanoinjection. Major congratulations to first author Katrin Mangold, PhD student in the lab. The developing mouse embryo is poorly amenable to genetic manipulations that are typically needed for functional gene analysis studies. In this issue, Mangold et al. develop NEPTUNE, a method to rapidly and flexibly transduce the neural plate with gene-expression-modifying viruses prior to neurulation to target the future adult nervous system. The cover image draws parallels between Neptune “the blue planet” in our solar system, Neptune the Roman god of the sea and freshwater, and NEPTUNE the method in which amniotic fluid is injected to control cell-type-specific expression and functional outcomes in brain and spinal cord. Artwork by illustrator Mattias Karlen, cover concept by E.R. Andersson.

June 2021
We welcome our new postdoc Elisabeth Verboven, who performed her PhD in the Georg Halder Lab at VIB – KU Leuven, Belgium.

We welcome our new postdoc Lenka Belicova, who performed her PhD in the Marino Zerial Lab at the Max Planck Institute of Molecular Cell Biology and Genetics, Germany.
May 2021
ERA Lab contributes to consensus paper on nomenclature for organoids, as HPB Consortium member: Marsee et al, Cell Stem Cell 2021.
Our analysis of gender bias in review processes is out: Andersson et al Front. Res. Metr. Anal. 2021. We find that among the highest-ranked applicants, women with equal quantifiable merits are ranked lower than men with equal merits. Fun anecdote: another high-impact journal rejected this paper, after revisions, because reviewer X “didn’t believe gender bias exists”.
April 2021
Our collaborative paper with Igor Adameyko lab is out in Nature Genetics! Polina Kameneva, Artem V. Artemov and colleagues used multiple approaches to lineage trace neural crest cells and identify sympathoblast lineages of relevance to neuroblastoma. We contributed with in utero transduction and lentiviral barcode lineage tracing of neural crest (Fig 4, see also NEPTUNE technique described in Mangold et al Cell Reports Methods 2021 above). This project was supported by, and is part of, our Knut and Alice Wallenberg project, a collaboration with Igor Adameyko, Jonas Frisén, Niklas Björkström and Ulrika Marklund groups.

March 2021
Two new arrivals!
We welcome our new postdoc Paulina Zydowicz-Machtel, who performed her PhD in the Jerzy Ciesiołka lab at the Institute of Bioorganic Chemistry, Polish Academy of Sciences, Poland.
We also welcome our new undergraduate student, Luisa Dehandschutter Gortari, from KU Leuven, Belgium.
Feb 2021
Two publications from our lab, out on the same day!
- Emma Andersson’s commentary in Science is out (full text link available under The Lab > Publications). In this commentary, Emma discusses two exciting new papers in Science investigating hepatocyte homeostasis and regeneration after injury.
- Our DUCT paper is out in eLife! Simona Hankeova, Jakub Salplachta and colleagues developed a technique to quantify the 3D architecture of the biliary and vascular trees in wild type and Alagille syndrome mice. We show that recovery occurs with distinct architectural mechanisms in hilar and peripheral liver, with resultant architecture that could be severely misinterpreted in 2D sections.
2020
Sept 30, 2020
Andersson lab is awarded a Knut and Alice Wallenberg project grant to study lineages and fate choices accross organ systems during development. This major project grant is a collaboration with Igor Adameyko, Niklas Björkström, Jonas Frisén and Ulrika Marklund. We are humbled and grateful to the KAW Foundation and the reviewers – and so excited to do this project!

Jan 2020
Andersson Lab is awarded the Swedish Foundation’s Starting Grant – an equivalent to the European Research Council Starting Grant. This collaboration between the Erling-Persson Family Foundation, the Kempe Foundations, the Foundation Olle Engkvist Byggmästare, Ragnar Söderberg Foundation and Riksbankens Jubileumsfond welcomes applications from researchers ranked A in the ERC Starting Grant, and recommended for funding, but for whom there was not sufficient funds in the ERC budget.
Read the press release here, and an interview here. We are deeply grateful to all the participating foundations for their initiative, in particular the Ragnar Söderberg Foundation that funded our Swedish Foundation Starting Grant in this round.
For more events, please see our News Archive.